|
Computational Structural Biology Group Head: Gunnar F. Schröder Institute of Biological Information Processing (IBI-7) Forschungszentrum Jülich and Professor of Molecular Biophysics at Heinrich-Heine University Düsseldorf |
|
|
The group uses cryo-electron microscopy (cryo-EM) to determine the structure of amyloid proteins. We aim to understand the molecular basis of diseases that involve amyloid aggregation, such as Alzheimer's, Parkinsons's, and diabetes. For this, we study the polymorphism of amyloid fibrils of Aβ, α-synuclein, IAPP, and amyloid model systems such as SH3.
Furthermore, the group develops computational methods for modeling and refinement of protein structures using low resolution or sparse experimental data. The focus is on the integration of structure prediction approaches with experimental data from different sources such as single particle cryo-EM, X-ray crystallography and NMR. |
|
| We are working on the prediction of local structural details using molecular dynamics simulation techniques to improve low-resolution model building and homology modeling. The goal is to reliably describe the structure and conformational dynamics of proteins from low or intermediate resolution data. | |
News
|
New publications
Check out our new paper on mouse model amyloid fibril structures by cryo-EM in Nature Neuroscience: 
Cryo-EM of Aβ fibrils from mouse models find tg-APPArcSwe fibrils resemble those found in patients with sporadic Alzheimer’s disease . Mara Zielinski*, Fernanda S. Peralta Reyes*, Lothar Gremer, Sarah Schemmert, Benedikt Frieg, Luisa U. Schäfer, Antje Willuweit, Lili Donner, Margitta Elvers, Lars N. G. Nilsson, Stina Syvänen, Dag Sehlin, Martin Ingelsson, Dieter Willbold and Gunnar F. Schröder Nature Neuroscience Volume 26 | December 2023 | 2073–2080 Our paper on Cryo-EM structures of lipidic fibrils of amyloid-β (1-40) by Benedikt Frieg and Mookyoung Han has just been accepted in Nature Communications 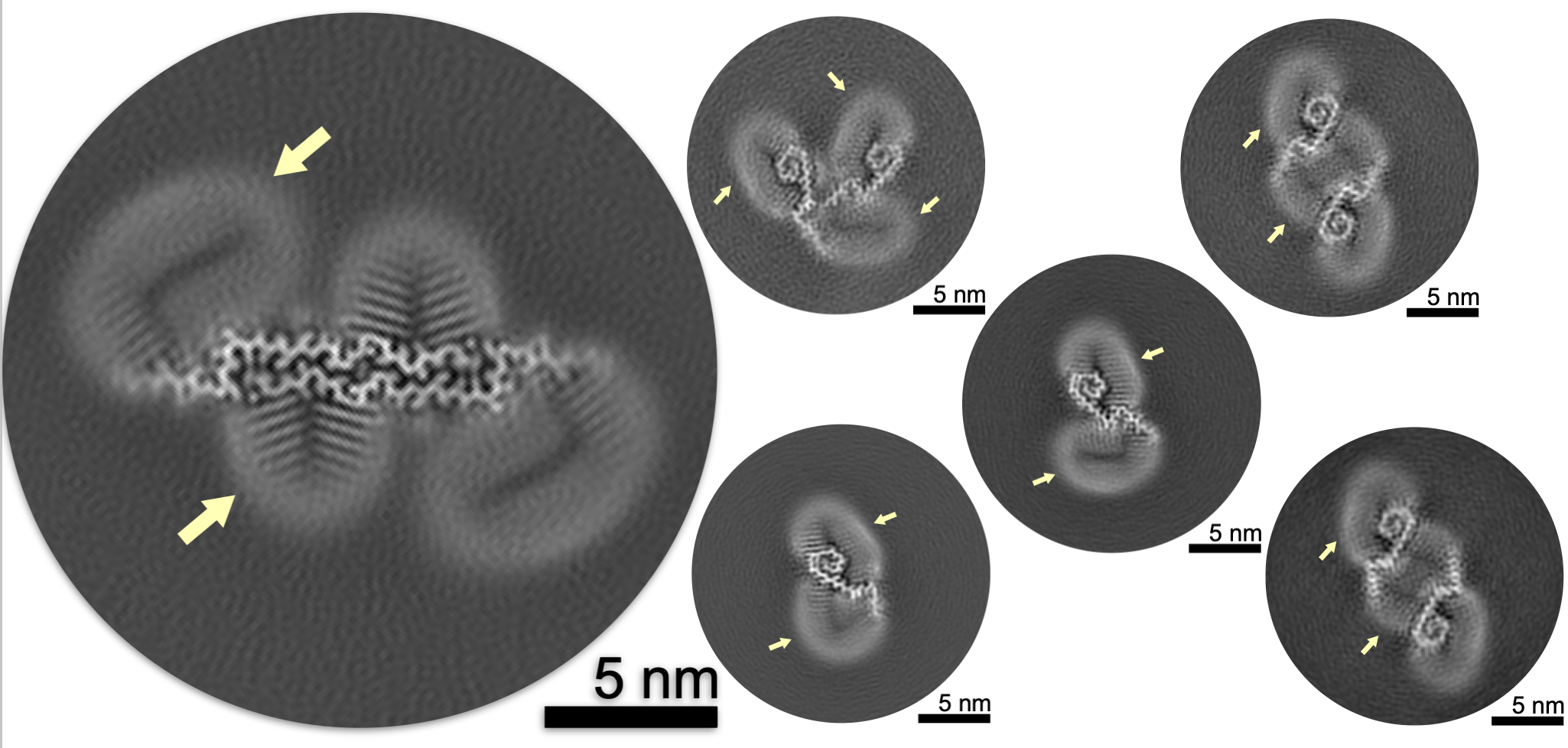
|